Publications
| 57. | Knisch, A.; Falcone, B.N.; Hirst J.D., Practical guidelines for optimising free energy calculations using thermodynamic integration. Chem. Phys. Lett., 880, 142395 (2025). DOI: http://dx.doi.org/10.1016/j.cplett.2025.142395 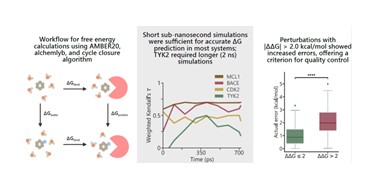 |
| 56. | Liu, Y.; Hirst, J.D.; Ren, J.; Tang, B.; Towey, D., Chemically-aware Attention-based Multi-modal Fusion Framework for Molecular Representation Learning. Proc. 49th IEEE Intl. Conf. Computers, Software, and Applications (COMPSAC 2025), Toronto, Canada, 1828–1833 (2025). DOI: http://dx.doi.org/10.1109/COMPSAC65507.2025.00250 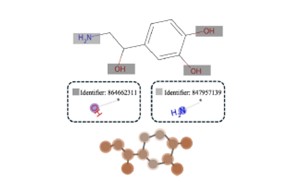 |
| 55. | Blackshaw, T.M.; Davies, J.C.; Spoerer, K.T.; Hirst, J.D., Enhancing Monte Carlo tree search for retrosynthesis. J. Chem. Inf. Model., 65, 6537–6546 (2025). DOI: http://dx.doi.org/10.1021/acs.jcim.5c00417 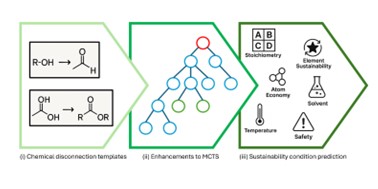 |
| 54. | Nwafor, P.; Gurung, S.; van Krimpen, P.; Schnaubert, L.; Jolley, K.; Pearman-Kanza, S.; Willoughby, C.; Hirst, J.D., AI4Green4Students: Promoting sustainable chemistry in undergraduate laboratories with an electronic lab notebook. J. Chem. Ed., 102, 2720–2731 (2025). DOI: http://dx.doi.org/doi.org/10.1021/acs.jchemed.4c01393 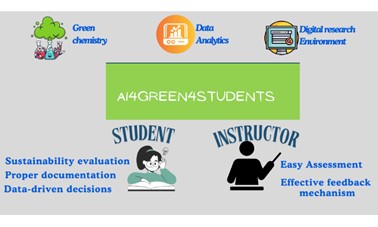 |
| 53. | Boobier, S.; Heeley, J.; Gärtner, T.; Hirst, J.D., Interactive Knowledge-based Kernel PCA for Green Solvent Selection. ACS Sus. Chem. Eng., 13, 4349–4368 (2025). DOI: http://dx.doi.org/10.1021/acssuschemeng.4c07974  |
| 52. | Liu, Y.; Duo, L.; Hirst, J.D.; Ren, J.; Tang, B.; Towey, D., Three-branch Molecular Representation Learning Framework for Predicting Molecular Properties in Drug Discovery. Proc. 48th IEEE Intl. Conf. Computers, Software, and Applications (COMPSAC 2024), Osaka, Japan, 1988–1994 (2024). DOI: http://dx.doi.org/10.1109/COMPSAC61105.2024.00317  |
| 51. | Davies, J.C.; Hirst, J.D., Software tools for green and sustainable chemistry. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering, Ed. Béla Török, ISBN 9780124095472 (2024). DOI: http://dx.doi.org/10.1016/B978-0-443-15742-4.00049-1  |
| 50. | Duo, L.; Liu, Y.; Ren, J.; Tang, B.; Hirst, J.D., Artificial Intelligence for Small Molecule Anti-Cancer Drug Discovery. Expert Opin. Drug Discov., 19, 933–948 (2024). DOI: http://dx.doi.org/10.1080/17460441.2024.2367014  |
| 49. | Heeley, J.; Boobier, S.; Hirst, J.D., Solvent Flashcards: A Visualisation Tool for Sustainable Chemistry. J. Cheminf., 16, 60 (2024). DOI: http://dx.doi.org/10.1186/s13321-024-00854-9  |
| 48. | Cooley, I.; Boobier, S.; Hirst, J.D.; Besley, E., Predicting biogas separation in metal organic frameworks. Commun. Chem., 7, 102 (2024). DOI: http://dx.doi.org/10.1038/s42004-024-01166-7  |
| 47. | Duo, L.; Liu, Q.; Chen, Y.; Low, S.S.; Ren, J.; Hirst, J.D.; Xie, H.; Tang, B., Discovery of Novel SOS1 Inhibitors using Machine Learning. RSC Med. Chem., 15, 1392–1403 (2024). DOI: http://dx.doi.org/10.1039/D4MD00063C  |
| 46. | Hirst, J.D., Boobier, S., Coughlan, J., Streets, J., Jacob, P., Pugh, O., Ozcan, E., Woodward, S., ML meets MLn: machine learning concepts in ligand promoted homogeneous catalysis. Artificial Intelligence Chem., 1, 100006 (2023). DOI: http://dx.doi.org/10.1016/j.aichem.2023.100006  |
| 45. | Chio, H.; Guest, E.E.; Hobman, J.L.; Dottorini, T.; Hirst, J.D. & Stekel, D.J., Predicting bioactivity of antibiotic metabolites by molecular docking. J. Mol. Graph. Model., 123, 108508 (2023). DOI: http://dx.doi.org/10.1016/j.jmgm.2023.108508  |
| 44. | Boobier, S., Davies, J.C., Derbenev, I.N., Handley, C.M. & Hirst, J.D., AI4Green: An Open-Source ELN for Green and Sustainable Chemistry. J. Chem. Inf. Mod., 63, 2895–2901 (2023). DOI: http://dx.doi.org/10.1021/acs.jcim.3c00306  |
| 43. | Redshaw, J.; Ting, D.S.J.; Brown, A.; Gaertner, T.; Hirst, J.D., Krein Support Vector Machine Classification of Antimicrobial Peptides. Digital Discovery, 2, 502–511 (2023). DOI: http://dx.doi.org/10.1039/D3DD00004D  |
| 42. | Silva, A.F.; Guest, E.E.; Falcone, B.N.; Pickett, S.D.; Rogers, D.M.; Hirst, J.D., Free energy perturbation calculations of tetrahydroquinolines complexed to the first bromodomain of BRD4. Mol. Phys., 121, e2124201 (2023). DOI: http://dx.doi.org/10.1080/00268976.2022.2124201  |
| 41. | Davies, J.C.; Pattison, D. & Hirst, J.D., Machine learning for yield prediction for chemical reactions using in situ sensors. J. Mol. Graph. Mod., 118, 108356 (2023). DOI: http://dx.doi.org/10.1016/j.jmgm.2022.108356  |
| 40. | Haywood, A.L.; Redshaw, J.; Hanson-Heine, M.W.D.; Taylor, A.; Brown, A.; Mason, A.M.; Gaertner, T. and Hirst, J.D., Kernel methods for predicting yields of chemical reactions. J. Chem. Inf. Mod., 62, 2077–2091 (2022). DOI: http://dx.doi.org/10.1021/acs.jcim.1c00699  |
| 39. | Guest, E.E., Cervantes Vasquez, L.F., Pickett, S.D., Brooks, C.L., Hirst, J.D., Alchemical free energy methods applied to complexes of the first bromodomain of BRD4. J. Chem. Inf. Mod., 62, 1458–1470 (2022). DOI: http://dx.doi.org/10.1021/acs.jcim.1c01229  |
| 38. | Derbenev, I.N.; Dowden, J.; Twycross, J.; Hirst, J.D., Software tools for green and sustainable chemistry. Curr. Opin. Green Sus. Chem., 35, 100623 (2022). DOI: http://dx.doi.org/10.1016/j.cogsc.2022.100623  |
| 37. | Zhou, J.; Wu, S.; Lee, B.G.; Chen, T.; He, Z.; Lei, Y.; Tang, B.; Hirst, J.D., Machine learning enabled virtual screening for inhibitors of lysine-specific histone demethylase 1. Molecules, 26, 7492 (2021). DOI: http://dx.doi.org/10.3390/molecules26247492  |
| 36. | Guest, E.E.; Pickett, S.D.; Hirst, J.D., Structural variation of protein-ligand complexes of the first bromodomain of BRD4. Org. Bio. Chem., 19, 5632–5641 (2021). DOI: http://dx.doi.org/10.1039/D1OB00658D  |
| 35. | Guest, E.E., Oatley, S.A., Macdonald, S.J.M., Hirst, J.D., Molecular simulation of aVb6 integrin inhibitors. J. Chem. Inf. Mod., 60, 5487–5498 (2020). DOI: http://dx.doi.org/10.1021/acs.jcim.0c00254  |
| 34. | Robinson H; Oatley SA; Rowedder JE; Slade P; Macdonald SJF; Hirst JD; McInally T & Moody CJ., Late stage functionalization via Chan-Lam amination: rapid access to potent and selective integrin inhibitors. Chem. Eur. J., 26, 7678–7684 (2020). DOI: http://dx.doi.org/10.1002/chem.202001059 |
| 33. | Haywood, A.L., Redshaw, J., Gaertner, T., Taylor, A., Mason, A.M. & Hirst, J.D., Machine Learning for Chemical Synthesis. In Machine Learning in Chemistry: The Impact of Artificial Intelligence, Ed. Cartwright, H. RSC, London, 169–194 (2020). DOI: http://dx.doi.org/10.1039/9781839160233-00169 |
| 32. | Oglic, D., Oatley, S.A., Macdonald, S.J.F., McInally, T., Garnett, R., Hirst, J.D. & Gärtner, T., Active search for computer-aided drug design. Mol. Inf., 37, 1700130 (2018). DOI: http://dx.doi.org/10.1002/minf.201700130 |
| 31. | Shaw, D.J., Hill, R.E., Simpson, N., Husseini, F.S., Robb, K., Greetham, G.M., Towrie, M., Parker, A.W., Robinson, D., Hirst, J.D., Hoskisson, P.A. & Hunt, N.T., Examining the role of protein structural dynamics in drug resistance in Mycobacterium tuberculosis. Chem. Sci., 8, 8384–8399 (2017). DOI: http://dx.doi.org/10.1039/c7sc03336b |
| 30. | Mulholland, S., Turpin, E.R., Bonev, B.B. & Hirst, J.D. , Docking and molecular dynamics simulations of the ternary complex nisin2:lipid II. Sci. Rep, 6, 21185 (2016). DOI: http://dx.doi.org/10.1038/srep21185 |
| 29. | Aguado-Ullate, S., Baker, J.A., Gonzàlez-Gonzàlez, V., Müller, C., Hirst, J.D. & Carbó, J.J., A theoretical study of the activity in Rh-catalysed hydroformylation: the origin of the enhanced activity of the p-acceptor phosphinine ligand. Catal. Sci. Technol., 4, 979–987 (2014). DOI: http://dx.doi.org/10.1039/c3cy00956d |
| 28. | Turpin, E.R., Fang, H.-J., Thomas, N.R. & Hirst, J.D., Cooperativity and site selectivity in the ileal lipid binding protein. Biochemistry, 52, 4723–4733 (2013). DOI: http://dx.doi.org/10.1021/bi400192w |
| 27. | Hussain, A., Shaw, P.E. & Hirst, J.D., Molecular dynamics simulations and in silico peptide ligand screening of the Elk-1 ETS domain. J. Cheminf., 3, 49 (2011). DOI: http://dx.doi.org/10.1186/1758-2946-3-49 |
| 26. | Turpin, E.R., Bonev, B.B., Hirst, J.D., Stereoselective disulfide formation stabilizes the local peptide conformation in Nisin mimics. Biochemistry, 49, 9594–9603 (2010). DOI: http://dx.doi.org/10.1021/bi101214t |
| 25. | Hussain, A., Melville, J.L. & Hirst, J.D., Molecular docking and QSAR of aplyronine A and analogues: potent inhibitors of actin. J. Comput.-Aided Mol. Des., 24, 42005 (2010). DOI: http://dx.doi.org/10.1007/s10822-009-9307-y |
| 24. | Spowage, B.M., Bruce, C.L. & Hirst, J.D., Interpretable correlation descriptors for quantitative structure-activity relationships. J. Cheminf, 1, 22 (2009). DOI: http://dx.doi.org/10.1186/1758-2946-1-22  |
| 23. | Melville, J.L., Burke, E.K. & Hirst, J.D., Machine Learning in Virtual Screening. Comb. Chem. & High Thr. Scr., 12, 332–343 (2009). DOI: http://dx.doi.org/10.2174/138620709788167980 |
| 22. | Melville, J.L., Moal, I.H., Baker-Glenn, C., Shaw, P.E, Pattenden, G. & Hirst, J.D., The Structural Determinants of Macrolide-Actin Binding: In Silico Insights. Biophys. J., 92, 3862–3867 (2007). DOI: http://dx.doi.org/10.1529/biophysj.106.103580 |
| 21. | Melville, J.L. & Hirst, J.D., TMACC: Interpretable Correlation Descriptors for Quantitative Structure-Activity Relationships. J. Chem. Inf. Mod., 47, 626–634 (2007). DOI: http://dx.doi.org/10.1021/ci6004178  |
| 20. | Vincent, E., Saxton, J., Baker-Glenn, C., Moal, I., Hirst, J.D., Pattenden, G. & Shaw, P.E., Disruption of actin dynamics and SRF-dependent gene regulation by the marine macrolide ulapualide A and its synthetic analogues. Cell. Mol. Life Sci., 64, 487–497 (2007). DOI: http://dx.doi.org/10.1007/s00018-007-6427-1 |
| 19. | Dryden, I.L., Hirst, J.D. & Melville, J.L., Statistical analysis of unlabelled points: comparing molecules in cheminformatics. Biometrics, 63, 237–251 (2007). DOI: http://dx.doi.org/10.1111/j.1541-0420.2006.00622.x |
| 18. | Bruce, C.L., Melville, J.L., Pickett, S.D & Hirst, J.D., Contemporary QSAR Classifiers Compared. J. Chem. Inf. Mod., 47, 219–227 (2007). DOI: http://dx.doi.org/10.1021/ci600332j  |
| 17. | Melville, J.L., Riley, J.F. & Hirst, J.D., Similarity by Compression. J. Chem. Inf. Mod., 47, 25–33 (2007). DOI: http://dx.doi.org/10.1021/ci600384z  |
| 16. | Melville, J.L., Lovelock, K.J.R., Wilson, C., Allbutt, B., Burke, E.K., Lygo, B. & Hirst, J.D., Exploring Phase-Transfer Catalysis with Molecular Dynamics and 3D/4D Quantitative Structure-Selectivity Relationships. J. Chem. Inf. Mod., 45, 971–981 (2005). DOI: http://dx.doi.org/10.1021/ci050051l |
| 15. | McNeany, T.J. & Hirst, J.D., Inhibition of the Tyrosine Kinase, Syk, Analyzed by Stepwise Non-Parametric Regression. J. Chem. Inf. Mod., 45, 768–776 (2005). DOI: http://dx.doi.org/10.1021/ci049631t |
| 14. | Lygo, B., Andrews, B.I., Hirst, J.D., Melville, J.L., Peterson, J.A. & Slack, D., Rapid screening of cinchona alkaloid derived phase-transfer catalysts. Application in the optimization of a glycine imine alkylation. Chim Oggi, 22(9), 41920 (2004). DOI: http://dx.doi.org/10.1002/chin.200549258 |
| 13. | Melville, J.L. & Hirst, J.D., On the stability of CoMFA models. J. Chem. Inf. Comput. Sci., 44, 1294–1300 (2004). DOI: http://dx.doi.org/10.1021/ci049944o |
| 12. | Melville, J.L., Andrews, B.I., Lygo, B. & Hirst, J.D., Computational screening of combinatorial catalyst libraries. Chem. Comm., 12, 1410–1411 (2004). DOI: http://dx.doi.org/10.1039/b402378a  |
| 11. | Hirst, J.D., McNeany, T.J., Howe, T. & Whitehead, L., Application of Non- Parametric Regression to Quantitative Structure-Activity Relationships. Bioorg. Med. Chem., 10, 1037–1041 (2002). DOI: http://dx.doi.org/10.1016/S0968-0896(01)00359-5  |
| 10. | Constans, P & Hirst, J.D., Non-Parametric Regressors Applied to Quantitative Structure-Activity Relationships. J. Chem. Inf. Comput. Sci., 40, 452–459 (2000). DOI: http://dx.doi.org/10.1021/ci990082e |
| 9. | Hirst, J.D., Predicting Ligand Binding Energies. Curr. Opin. Drug Discovery & Development, 1, 28–33 (1998). |
| 8. | Vieth, M., Hirst, J.D., Dominy, B.N., Daigler, H. & Brooks III, C.L., Assessing Search Strategies for Flexible Docking. J. Comp. Chem., 19, 1623–1631 (1998). DOI: http://dx.doi.org/10.1002/(SICI)1096-987X(19981115)19:14<1623::AID-JCC8>3.0.CO;2-L |
| 7. | Vieth, M., Hirst, J.D., Kolinski, A. & Brooks III, C.L., Assessing Energy Functions for Flexible Docking. J. Comp. Chem., 19, 1612–1622 (1998). DOI: http://dx.doi.org/10.1002/(SICI)1096-987X(19981115)19:14<1612::AID-JCC7>3.0.CO;2-M |
| 6. | Vieth, M., Hirst, J.D. & Brooks III, C.L., Do Active Site Conformations of Small Ligands Correspond to Low Free Energy Structures? J. Comput.-Aided Mol. Des., 12, 563–572 (1998). DOI: http://dx.doi.org/10.1023/A:1008055202136 |
| 5. | Hirst, J.D., Nonlinear Quantitative Structure-Activity Relationship for the Inhibition of Dihydrofolate Reductase by Pyrimidines. J. Med. Chem., 39, 3526–3532 (1996). DOI: http://dx.doi.org/10.1021/jm960197z |
| 4. | King, R.D., Hirst, J.D. & Sternberg, M.J.E., Drug Design by Machine Learning: A Comparative Study. Applications in Artificial Intelligence, 9, 213–233 (1995). |
| 3. | Hirst, J.D., King, R.D. & Sternberg, M.J.E., Quantitative Structure-Activity Relationships by Neural Networks and Inductive Logic Programming II. The Inhibition of Dihydrofolate Reductase by Triazines. J. Comput.-Aided Mol. Des., 8, 421–432 (1994). DOI: http://dx.doi.org/10.1007/BF00125376 |
| 2. | Hirst, J.D., King, R.D. & Sternberg, M.J.E., Quantitative Structure-Activity Relationships by Neural Networks and Inductive Logic Programming I. The Inhibition of Dihydrofolate Reductase by Pyrimidines. J. Comput.-Aided Mol. Des., 8, 405–420 (1994). DOI: http://dx.doi.org/10.1007/BF00125375 |
| 1. | King, R.D., Hirst, J.D. & Sternberg, M.J.E., New Approaches to QSAR: Neural Networks and Machine Learning. Perspectives in Drug Discovery and Design, 1, 279–290 (1993). DOI: http://dx.doi.org/10.1007/BF02174529 |


