Publications
| 192. | Duo, L.; Tang, B.; Hirst, J.D.; Xie, H.; Ren, J., DMPE: A Dual-branch Molecular Property Encapsulation Framework with Kolmogorov-Arnold Networks. J. Comput. Chem., 46, e70273 (2025). DOI: http://dx.doi.org/10.1002/jcc.70273 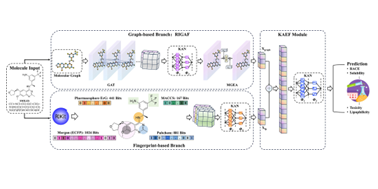 |
| 191. | Knisch, A.; Falcone, B.N.; Hirst J.D., Practical guidelines for optimising free energy calculations using thermodynamic integration. Chem. Phys. Lett., 880, 142395 (2025). DOI: http://dx.doi.org/10.1016/j.cplett.2025.142395 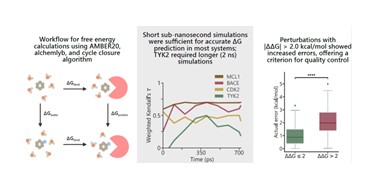 |
| 190. | Liu, Y.; Hirst, J.D.; Ren, J.; Tang, B.; Towey, D., Chemically-aware Attention-based Multi-modal Fusion Framework for Molecular Representation Learning. Proc. 49th IEEE Intl. Conf. Computers, Software, and Applications (COMPSAC 2025), Toronto, Canada, 1828–1833 (2025). DOI: http://dx.doi.org/10.1109/COMPSAC65507.2025.00250 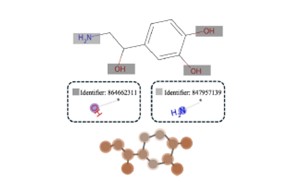 |
| 189. | Chaudhuri, S.; Huynh, B.C.; Amory, R.; Rogers, D.M.; Cranney, A.; Martínez-Martínez, L.A.; Macrì, T.; Jones, G.; Sheridan, E.; Khedkar, A.; Mineh, L.; Inzani, K.; Hirst J.D., Challenges and Advances in the Simulation of Targeted Covalent Inhibitors Using Quantum Computing. J. Phys. Chem. Lett., 16, 8536−8545 (2025). DOI: http://dx.doi.org/10.1021/acs.jpclett.5c01680  |
| 188. | Jiang, L.; Hirst, J.D.; Do, H., Interconnected morphological and electronic structure properties of the PBDB-T-2F (PM6) electron donor in bulk organic semiconductors. ACS Appl. Energ. Mat., 8, 11342–11352 (2025). DOI: http://dx.doi.org/10.1021/acsaem.5c01492  |
| 187. | Blackshaw, T.M.; Davies, J.C.; Spoerer, K.T.; Hirst, J.D., Enhancing Monte Carlo tree search for retrosynthesis. J. Chem. Inf. Model., 65, 6537–6546 (2025). DOI: http://dx.doi.org/10.1021/acs.jcim.5c00417 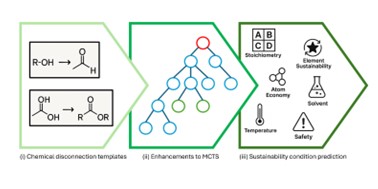 |
| 186. | Nwafor, P.; Gurung, S.; van Krimpen, P.; Schnaubert, L.; Jolley, K.; Pearman-Kanza, S.; Willoughby, C.; Hirst, J.D., AI4Green4Students: Promoting sustainable chemistry in undergraduate laboratories with an electronic lab notebook. J. Chem. Ed., 102, 2720–2731 (2025). DOI: http://dx.doi.org/doi.org/10.1021/acs.jchemed.4c01393  |
| 185. | Boobier, S.; Heeley, J.; Gärtner, T.; Hirst, J.D., Interactive Knowledge-based Kernel PCA for Green Solvent Selection. ACS Sus. Chem. Eng., 13, 4349–4368 (2025). DOI: http://dx.doi.org/10.1021/acssuschemeng.4c07974 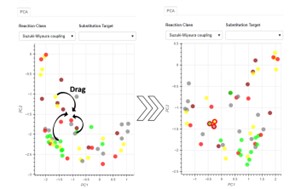 |
| 184. | Guo, Z.; Zhu, H.; Yan, Z.; Lei, L.; Wang, D.; Xi, Z.; Lian, Y.; Yu, J.; Fow, K.L.; Do, H.; Hirst, J.D.; Xu, M.; Wu, T., Manipulating Adsorbed Hydrogen on Lanthanum-Modified CuO: Industrial-Current-Density CO2 Electroreduction to C2+ products or CH4. Appl. Catal. B., 364, 124839 (2025). DOI: http://dx.doi.org/10.1016/j.apcatb.2024.124839 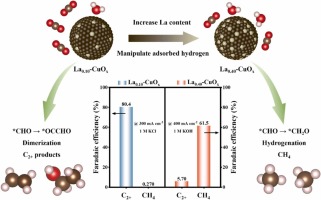 |
| 183. | Chaudhuri, S.; Rogers, D.M.; Hayes, C.J.; Inzani, K.; Hirst, J.D., Quantum Chemical Characterization of Rotamerism in Thio-Michael Additions for Targeted Covalent Inhibitors. J. Chem. Inf. Mod., 64, 7687–7697 (2024). DOI: http://dx.doi.org/10.1021/acs.jcim.4c01379  |
| 182. | Wang, D.; Pattenden, G.; Fow, K.L.; Stocks, M.J.; Hirst, J.D.; Tang, B., Theoretical Study on the Biosynthesis of the Mandapamates: Mechanistic Insights Using Density Functional Theory. J. Org. Chem., 89, 12946−12956 (2024). DOI: http://dx.doi.org/10.1021/acs.joc.4c00859 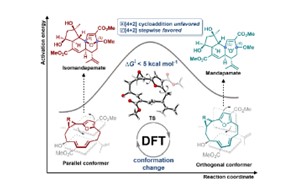 |
| 181. | Liu, Y.; Duo, L.; Hirst, J.D.; Ren, J.; Tang, B.; Towey, D., Three-branch Molecular Representation Learning Framework for Predicting Molecular Properties in Drug Discovery. Proc. 48th IEEE Intl. Conf. Computers, Software, and Applications (COMPSAC 2024), Osaka, Japan, 1988–1994 (2024). DOI: http://dx.doi.org/10.1109/COMPSAC61105.2024.00317 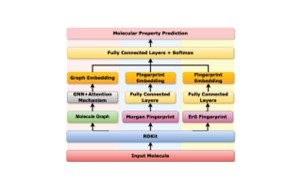 |
| 180. | Davies, J.C.; Hirst, J.D., Software tools for green and sustainable chemistry. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering, Ed. Béla Török, ISBN 9780124095472 (2024). DOI: http://dx.doi.org/10.1016/B978-0-443-15742-4.00049-1  |
| 179. | Duo, L.; Liu, Y.; Ren, J.; Tang, B.; Hirst, J.D., Artificial Intelligence for Small Molecule Anti-Cancer Drug Discovery. Expert Opin. Drug Discov., 19, 933–948 (2024). DOI: http://dx.doi.org/10.1080/17460441.2024.2367014 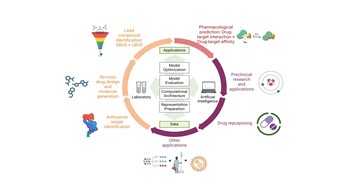 |
| 178. | Rogers, D.M.; Do, H.; Hirst, J.D., An Improved Diabatization Scheme for Computing the Electronic Circular Dichroism of Proteins. J. Phys. Chem. B, 128, 7350–7361 (2024). DOI: http://dx.doi.org/10.1021/acs.jpcb.4c02582 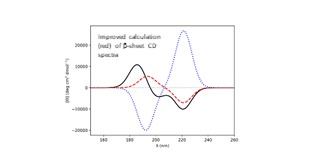 |
| 177. | Heeley, J.; Boobier, S.; Hirst, J.D., Solvent Flashcards: A Visualisation Tool for Sustainable Chemistry. J. Cheminf., 16, 60 (2024). DOI: http://dx.doi.org/10.1186/s13321-024-00854-9 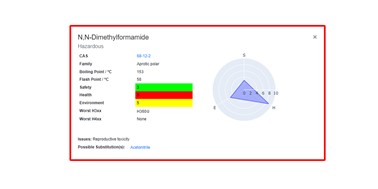 |
| 176. | Cooley, I.; Boobier, S.; Hirst, J.D.; Besley, E., Predicting biogas separation in metal organic frameworks. Commun. Chem., 7, 102 (2024). DOI: http://dx.doi.org/10.1038/s42004-024-01166-7 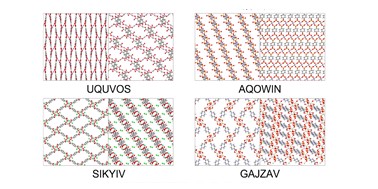 |
| 175. | Duo, L.; Liu, Q.; Chen, Y.; Low, S.S.; Ren, J.; Hirst, J.D.; Xie, H.; Tang, B., Discovery of Novel SOS1 Inhibitors using Machine Learning. RSC Med. Chem., 15, 1392–1403 (2024). DOI: http://dx.doi.org/10.1039/D4MD00063C 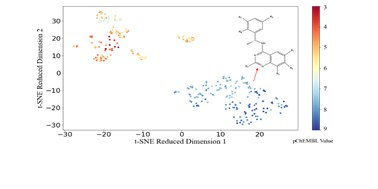 |
| 174. | Wang, D.; Zhou, T.; Ren, J.; Hirst, J.D.; Gao, Z.; Tang, B., Applications of quantum chemistry in biomimetic syntheses of polycyclic furanocembrane derivatives. Synlett., 35, 565–575 (2024). DOI: http://dx.doi.org/10.1055/a-2239-6577 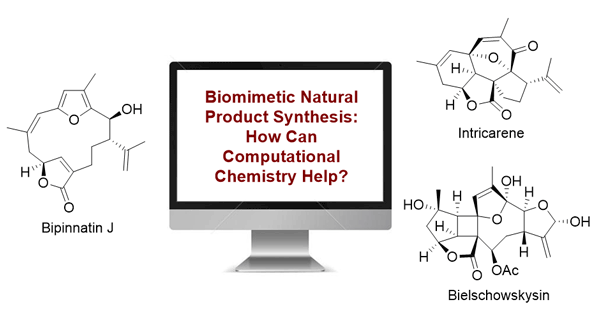 |
| 173. | Guo, Z.; Yang, F.; Li, X.; Zhu, H.; Do, H.; Fow, K.L.; Hirst, J. D.; Wu. W.; Ye, Q.; Peng, Y.; Wu, H. B.; Wu, A.; Xu, M., Electrocatalytic CO2 reduction to C2H4: From Lab to Fab. J. Energy Chem., 90, 540–564 (2024). DOI: http://dx.doi.org/10.1016/j.jechem.2023.11.019 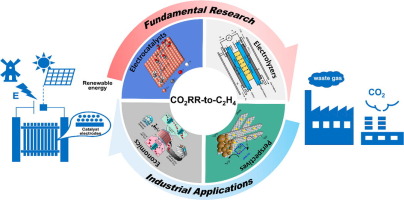 |
| 172. | King, T.E.; Laughton, C.A.; Humphrey, J.; Thomas, N.R.; Hirst, J.D., Optimising excipient properties to prevent aggregation in biopharmaceutical formulations. J. Chem. Inf. Mod., 64, 265−275 (2023). DOI: http://dx.doi.org/10.1021/acs.jcim.3c01898 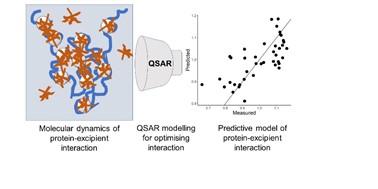 |
| 171. | Douaki, A.; Stuber, A.; Hengsteler, J.; Momotenko, D.; Rogers, D.M.; Hirst, J.D.; Nakatsuka, N.; Garoli, D., Theoretical visualization of divalent cations effects on aptamer recognition of neurotransmitter targets. Chem. Commun., 59, 14713–14716 (2023). DOI: http://dx.doi.org/10.1039/d3cc04334g  |
| 170. | Guo, Z.; Zhu, H.; Yang, G.; Wu, A.; Chen, Q.; Yan, Z.; Fow, K.L.; Do, H.; Hirst, J.D.; Wu, T.; Xu, M., Synergistic engineering of heteronuclear Ni-Ag dual-atom catalysts for high-efficiency CO2 electroreduction with nearly 100% CO selectivity. Chem. Eng. J., 476, 146556 (2023). DOI: http://dx.doi.org/10.1016/j.cej.2023.146556 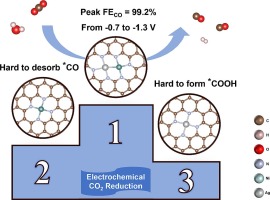 |
| 169. | Hirst, J.D., Boobier, S., Coughlan, J., Streets, J., Jacob, P., Pugh, O., Ozcan, E., Woodward, S., ML meets MLn: machine learning concepts in ligand promoted homogeneous catalysis. Artificial Intelligence Chem., 1, 100006 (2023). DOI: http://dx.doi.org/10.1016/j.aichem.2023.100006  |
| 168. | Chio, H.; Guest, E.E.; Hobman, J.L.; Dottorini, T.; Hirst, J.D. & Stekel, D.J., Predicting bioactivity of antibiotic metabolites by molecular docking. J. Mol. Graph. Model., 123, 108508 (2023). DOI: http://dx.doi.org/10.1016/j.jmgm.2023.108508  |
| 167. | Boobier, S., Davies, J.C., Derbenev, I.N., Handley, C.M. & Hirst, J.D., AI4Green: An Open-Source ELN for Green and Sustainable Chemistry. J. Chem. Inf. Mod., 63, 2895–2901 (2023). DOI: http://dx.doi.org/10.1021/acs.jcim.3c00306  |
| 166. | Redshaw, J.; Ting, D.S.J.; Brown, A.; Gaertner, T.; Hirst, J.D., Krein Support Vector Machine Classification of Antimicrobial Peptides. Digital Discovery, 2, 502–511 (2023). DOI: http://dx.doi.org/10.1039/D3DD00004D  |
| 165. | Jiang, L.; Hirst, J.D. & Do, H., Dynamic Disorder Drives Exciton Dynamics in Molecular Crystals: A Case Study. J. Phys. Chem. C, 127, 5519–5532 (2023). DOI: http://dx.doi.org/10.1021/acs.jpcc.2c07984  |
| 164. | Griffiths, R.C.; Smith, F.R.; Li, D.; Wyatt, J.; Rogers, D.M.; Long, J.E.; Cusin, L.M.L.; Tighe, P.J.; Layfield, R.; Hirst, J.D. & Mitchell, N.J. , Cysteine-Selective Modification of Peptides and Proteins via Desulfurative C-C Bond Formation. Chem. Eur. J., 29, e202202503 (2023). DOI: http://dx.doi.org/10.1002/chem.202202503  |
| 163. | Rogers, D.M.; Do, H. & Hirst, J.D., Electronic Circular Dichroism of Proteins Computed Using a Diabatisation Scheme. Mol. Phys., 121, e2133748 (2023). DOI: http://dx.doi.org/10.1080/00268976.2022.2133748  |
| 162. | Silva, A.F.; Guest, E.E.; Falcone, B.N.; Pickett, S.D.; Rogers, D.M.; Hirst, J.D., Free energy perturbation calculations of tetrahydroquinolines complexed to the first bromodomain of BRD4. Mol. Phys., 121, e2124201 (2023). DOI: http://dx.doi.org/10.1080/00268976.2022.2124201  |
| 161. | Davies, J.C.; Pattison, D. & Hirst, J.D., Machine learning for yield prediction for chemical reactions using in situ sensors. J. Mol. Graph. Mod., 118, 108356 (2023). DOI: http://dx.doi.org/10.1016/j.jmgm.2022.108356  |
| 160. | Lentz, J.C.; Cavanagh, R.; Moloney, C.; Falcone Pin, B.; Kortsen, K.; Fowler, H.; Jacob, P.L.; Krumins, E.; Clark, C.; Machado, F.; Breitkreuz, N.; Cale, B.; Goddard, A.R.; Hirst, J.D.; Taresco, V.; Howdle, S.M., N-Hydroxyethyl acrylamide as a functional eROP initiator for the preparation of nanoparticles in "greener" reaction conditions. Polymer Chem., 13, 6032–6045 (2022). DOI: http://dx.doi.org/10.1039/D2PY00849A  |
| 159. | Jiang, L.; Hirst, J.D. & Do, H., Structure-Property Relationships in Amorphous Thieno[3,2-b]thiophene- Diketopyrrolopyrrole-Thiophene-Containing Polymers. J. Phys. Chem. C, 126, 10842–10854 (2022). DOI: http://dx.doi.org/10.1021/acs.jpcc.2c01650  |
| 158. | Haywood, A.L.; Redshaw, J.; Hanson-Heine, M.W.D.; Taylor, A.; Brown, A.; Mason, A.M.; Gaertner, T. and Hirst, J.D., Kernel methods for predicting yields of chemical reactions. J. Chem. Inf. Mod., 62, 2077–2091 (2022). DOI: http://dx.doi.org/10.1021/acs.jcim.1c00699  |
| 157. | Guest, E.E., Cervantes Vasquez, L.F., Pickett, S.D., Brooks, C.L., Hirst, J.D., Alchemical free energy methods applied to complexes of the first bromodomain of BRD4. J. Chem. Inf. Mod., 62, 1458–1470 (2022). DOI: http://dx.doi.org/10.1021/acs.jcim.1c01229  |
| 156. | Derbenev, I.N.; Dowden, J.; Twycross, J.; Hirst, J.D., Software tools for green and sustainable chemistry. Curr. Opin. Green Sus. Chem., 35, 100623 (2022). DOI: http://dx.doi.org/10.1016/j.cogsc.2022.100623  |
| 155. | Gaughan, S.J.H., Hirst, J.D., Croft, A.K. & Jaeger, C.M., The effect of oriented electric fields on biologically relevant iron-sulfur clusters: Tuning redox reactivity for catalysis. J. Chem. Inf. Mod., 62, 591–601 (2022). DOI: http://dx.doi.org/10.1021/acs.jcim.1c00791  |
| 154. | Zhou, J.; Wu, S.; Lee, B.G.; Chen, T.; He, Z.; Lei, Y.; Tang, B.; Hirst, J.D., Machine learning enabled virtual screening for inhibitors of lysine-specific histone demethylase 1. Molecules, 26, 7492 (2021). DOI: http://dx.doi.org/10.3390/molecules26247492  |
| 153. | Guest, E.E.; Pickett, S.D.; Hirst, J.D., Structural variation of protein-ligand complexes of the first bromodomain of BRD4. Org. Bio. Chem., 19, 5632–5641 (2021). DOI: http://dx.doi.org/10.1039/D1OB00658D  |
| 152. | Robinson, D., Alarfaji, S. & Hirst, J.D., Benzene and mono-substituted derivatives: diabatic nature of the oscillator strengths of S1 <-- S0 transitions. . J. Phys. Chem. A, 125, 5237–5245 (2021). DOI: http://dx.doi.org/10.1021/acs.jpca.1c01685  |
| 151. | Pacheco, A.A. C.; da Silva Filho, A.F.; Kortsen, K.; Hanson-Heine, M.W.D.; Taresco, V.; Hirst, J.D.; Lansalot, M.; D'Agosto, F. & Howdle, S.M., Influence of structure and solubility of chain transfer agents on the RAFT controlled dispersion polymerisation in scCO2. Chem. Sci., 12, 1016–1030 (2021). DOI: http://dx.doi.org/10.1039/D0SC05281G  |
| 150. | Segatta, F., Rogers, D.M., Dyer, N.T., Guest, E.E., Li, Z., Do, H., Nenov, A., Garavelli, M., Hirst, J.D., Near-ultraviolet circular dichroism and two-dimensional spectroscopy of polypeptides. Molecules, 26, 396 (2021). DOI: http://dx.doi.org/10.3390/molecules26020396 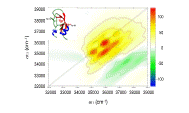 |
| 149. | Guest, E.E., Oatley, S.A., Macdonald, S.J.M., Hirst, J.D., Molecular simulation of aVb6 integrin inhibitors. J. Chem. Inf. Mod., 60, 5487–5498 (2020). DOI: http://dx.doi.org/10.1021/acs.jcim.0c00254  |
| 148. | Ye, S., Zhong, K., Zhang, J., Hu, W., Hirst, J.D., Zhang, G., Mukamel, S., Jiang, J., A transferable machine learning protocol for predicting protein amide-I infrared spectra. J. Am. Chem. Soc., 142, 19071–19077 (2020). DOI: http://dx.doi.org/10.1021/jacs.0c06530  |
| 147. | Sole-Daura, A.; Rodriguez-Fortea, A.; Poblet, J.M.; Robinson, D.; Hirst, J.D.; Carbo, J.J., Origin of selectivity in protein hydrolysis by Zr(IV)-containing metal oxides as artificial proteases. ACS Catalysis, 10, 13455–13467 (2020). DOI: http://dx.doi.org/10.1021/acscatal.0c02848  |
| 146. | Spankie, T.J., Haywood, A.L; Dottorini, T., Barrow, P.A. & Hirst, J.D., Interaction of the maturation protein of the bacteriophage MS2 and the sex pilus of the Escherichia coli F plasmid. J. Mol. Graph. Mod., 101, 107723 (2020). DOI: http://dx.doi.org/10.1016/j.jmgm.2020.107723  |
| 145. | Li, Z. & Hirst, J.D., Computed optical spectra of SARS-CoV-2 proteins. Chem. Phys. Lett., 758, 137935 (2020). DOI: http://dx.doi.org/10.1016/j.cplett.2020.137935  |
| 144. | Baiz, C.R.; Blasiak, B.; Bredenbeck, J.; Cho, M.; Choi, J.-H.; Corcelli, S.A.; Dijkstra, A.G.; Feng, C.J.; Garrett-Roe, S.; Nien-Hui Ge, N.-H.; Hanson-Heine, M.W.D.; Hirst, J.D.; Jansen, T.L.C.; Kwac, K.; Kubarych, K.J.; Londergan, C.H.; Maekawa, H.; Reppert, M.; Saito, S.; Roy, S.; Skinner, J.L.; Stock, G.; Straub, J.E.; Thielges, M.C.; Tominaga, K.; Tokmakoff, A.; Torii, H.; Wang, L.; Webb, L.J.; Zanni, M.T., Vibrational Frequency Map, Vibrational Spectroscopy, and Intermolecular Interaction. . Chem. Rev., 120, 7152–7218 (2020). DOI: http://dx.doi.org/10.1021/acs.chemrev.9b00813  |
| 143. | Jiang, L., Rogers, D.M., Hirst, J.D. & Do, H., Force fields for macromolecular assemblies containing diketopyrrolopyrrole and thiophene. J. Chem. Theor. Comput., 16, 5150–5162 (2020). DOI: http://dx.doi.org/10.1021/acs.jctc.0c00399  |
| 142. | Hanson-Heine, M.W.D. & Hirst, J.D., Mobius and Huckel cyclacenes with Dewar and Ladenburg defects. J. Phys. Chem. A, 124, 5408–5414 (2020). DOI: http://dx.doi.org/10.1021/acs.jpca.0c04137  |
| 141. | Auvray, F. & Hirst, J.D., Unfolding dynamics of a photo-switchable helical peptide. J. Phys. Chem. B, 124, 5380–5392 (2020). DOI: http://dx.doi.org/10.1021/acs.jpcb.0c04017  |
| 140. | Robinson H; Oatley SA; Rowedder JE; Slade P; Macdonald SJF; Hirst JD; McInally T & Moody CJ., Late stage functionalization via Chan-Lam amination: rapid access to potent and selective integrin inhibitors. Chem. Eur. J., 26, 7678–7684 (2020). DOI: http://dx.doi.org/10.1002/chem.202001059 |
| 139. | Hanson-Heine, M.W.D., Rogers, D.M., Woodward, S. & Hirst, J.D. , Dewar benzene ground states found in cyclacene nanobelts. J. Phys. Chem. Lett., 11, 3769–3772 (2020). DOI: http://dx.doi.org/10.1021/acs.jpclett.0c01027 |
| 138. | Haywood, A.L., Redshaw, J., Gaertner, T., Taylor, A., Mason, A.M. & Hirst, J.D., Machine Learning for Chemical Synthesis. In Machine Learning in Chemistry: The Impact of Artificial Intelligence, Ed. Cartwright, H. RSC, London, 169–194 (2020). DOI: http://dx.doi.org/10.1039/9781839160233-00169 |
| 137. | Rogers, D.M, Jasim, S.B, Dyer, N.T, Auvray, F, Rèfrègiers, M, Hirst, J.D., Electronic circular dichroism of proteins. Chem, 5, 2751–2774 (2019). DOI: http://dx.doi.org/10.1016/j.chempr.2019.07.008 |
| 136. | Auvray, F, Dennetiere D, Giulianib A, Jamme F, Wien F, Polack F, Menneglier C, Lagarde B, Hirst JD, Rèfrègiers M., Time resolved transient circular dichroism spectroscopy based on synchrotron natural polarization. Stuct Dynamics, 6, 54307 (2019). DOI: http://dx.doi.org/10.1063/1.5120346 |
| 135. | Michaelis M, Hildebrand N, Meissner RH, Wurzler N, Li Z, Hirst JD, Micsonai A, Kardos J, Delle Piane M & Colombi Ciacchi L., Impact of the conformational variability of oligopeptides on the computational prediction of their CD spectra. J Phys Chem B, 123, 6694–6704 (2019). DOI: http://dx.doi.org/10.1021/acs.jpcb.9b03932 |
| 134. | Hildebrand, N., Michaelis, M., Wurzler, N., Li, Z., Hirst, J.D., Misconai, A., Kardoe, J., Koeppen, S., delle Piane, M., Bussi, G. & Ciacchi, L.C. , Atomistic details of protein conformational changes upon adsorption on silica. ACS Biomat. Sci. Eng., 4, 4036–4050 (2018). DOI: http://dx.doi.org/10.1021/acsbiomaterials.8b00819 |
| 133. | Jasim, S.B., Li, Z., Guest, E.E. & Hirst, J.D., DichroCalc: improvements in computing protein circular dichroism spectroscopy in the near-ultraviolet. J. Mol. Biol., 430, 2196–2202 (2018). DOI: http://dx.doi.org/10.1016/j.jmb.2017.12.009 |
| 132. | Oglic, D., Oatley, S.A., Macdonald, S.J.F., McInally, T., Garnett, R., Hirst, J.D. & Gärtner, T., Active search for computer-aided drug design. Mol. Inf., 37, 1700130 (2018). DOI: http://dx.doi.org/10.1002/minf.201700130 |
| 131. | Shaw, D.J., Hill, R.E., Simpson, N., Husseini, F.S., Robb, K., Greetham, G.M., Towrie, M., Parker, A.W., Robinson, D., Hirst, J.D., Hoskisson, P.A. & Hunt, N.T., Examining the role of protein structural dynamics in drug resistance in Mycobacterium tuberculosis. Chem. Sci., 8, 8384–8399 (2017). DOI: http://dx.doi.org/10.1039/c7sc03336b |
| 130. | Suess, C., Hirst, J.D. & Besley, N.A. , Modelling tryptophan-->heme electron and excitation energy transfer rates in myoglobin. J. Comput. Chem., 38, 1495–1502 (2017). DOI: http://dx.doi.org/10.1002/jcc.24793 |
| 129. | Li, Z. & Hirst, J.D., Vibrational structure in the near-ultraviolet electronic circular dichroism spectra of proteins. Chem. Sci., 8, 4318–4333 (2017). DOI: http://dx.doi.org/10.1039/C7SC00586E |
| 128. | Husseini, F.S., Robinson, D., Hunt, N.T., Parker, A.W. & Hirst, J.D. , Computing infrared spectra of proteins using the exciton model. J. Comput. Chem., 38, 1362–1375 (2017). DOI: http://dx.doi.org/10.1002/jcc.24674 |
| 127. | Solè-Daura, A., Goovaerts, V., Stroobants, K., Absillis, G., Jimènez-Lozano, P., Poblet, J.M., Hirst, J.D., Parac-Vogt, T.N. & Carbó, J.J., Probing polyoxometalate-protein interactions using molecular dynamics simulations. Chem. Eur. J., 22, 15280–15289 (2016). DOI: http://dx.doi.org/10.1002/chem.201602263 |
| 126. | Mulholland, S., Turpin, E.R., Bonev, B.B. & Hirst, J.D. , Docking and molecular dynamics simulations of the ternary complex nisin2:lipid II. Sci. Rep, 6, 21185 (2016). DOI: http://dx.doi.org/10.1038/srep21185 |
| 125. | Hanson-Heine, M.W.D., Husseini, F., Hirst, J.D. & Besley, N.A., Simulation of the two-dimensional infrared spectroscopy of peptides using localized normal modes. J. Chem. Theor. Comput., 12, 1905–1918 (2016). DOI: http://dx.doi.org/10.1021/acs.jctc.5b01198 |
| 124. | Li, Z., Robinson, D. & Hirst, J.D., Vibronic structure in the far-UV electronic circular dichroism spectra of proteins. Faraday Discussion, 177, 329–344 (2015). DOI: http://dx.doi.org/10.1039/C4FD00163J |
| 123. | Turpin, E.R., Mulholland, S., Bonev, B. & Hirst, J.D., New CHARMM force field parameters for dehydrated amino acid residues, the key to lantibiotic molecular dynamics simulations. RSC Advances., 4, 48621–48631 (2014). DOI: http://dx.doi.org/10.1039/c4ra09897h |
| 122. | Baker, J.A. & Hirst, J.D., Accelerating electrostatic pair methods on graphical processing units to study molecules in supercritical carbon dioxide. Faraday Discussion, 169, 343–357 (2014). DOI: http://dx.doi.org/10.1039/c4fd00012a |
| 121. | Hirst, J.D., Glowacki, D., Baaden, M., Molecular simulations and visualization: introduction and overview. Faraday Discussion, 169, 922 (2014). DOI: http://dx.doi.org/10.1039/c4fd90024c |
| 120. | Aguado-Ullate, S., Baker, J.A., Gonzàlez-Gonzàlez, V., Müller, C., Hirst, J.D. & Carbó, J.J., A theoretical study of the activity in Rh-catalysed hydroformylation: the origin of the enhanced activity of the p-acceptor phosphinine ligand. Catal. Sci. Technol., 4, 979–987 (2014). DOI: http://dx.doi.org/10.1039/c3cy00956d |
| 119. | Hill R.E., Hunt N.T. & Hirst J.D., Studying biomacromolecules with two-dimensional infrared spectroscopy. Adv. Prot. Chem. Str. Biol., 93, 13150 (2013). DOI: http://dx.doi.org/10.1016/B978-0-12-416596-0.00001-4 |
| 118. | Turpin, E.R., Fang, H.-J., Thomas, N.R. & Hirst, J.D., Cooperativity and site selectivity in the ileal lipid binding protein. Biochemistry, 52, 4723–4733 (2013). DOI: http://dx.doi.org/10.1021/bi400192w |
| 117. | Turpin, E.R. & Hirst, J.D., Transformation of the dihedral corrective map for D-amino residues using the CHARMM force field. Chem. Phys. Lett., 543, 142–147 (2012). DOI: http://dx.doi.org/10.1016/j.cplett.2012.06.041 |
| 116. | Do, H., Hirst, J.D. & Wheatley, R.J., Calculation of partition functions and free energies of a binary mixture using the energy partitioning method: application to CO2 + CH4. J. Phys. Chem. B, 116, 4535–4542 (2012). DOI: http://dx.doi.org/10.1021/jp212168f |
| 115. | Hussain, A., Shaw, P.E. & Hirst, J.D., Molecular dynamics simulations and in silico peptide ligand screening of the Elk-1 ETS domain. J. Cheminf., 3, 49 (2011). DOI: http://dx.doi.org/10.1186/1758-2946-3-49 |
| 114. | Do, H., Hirst, J.D. & Wheatley, R.J., Rapid calculation of the partition function of fluids. J. Chem. Phys., 135, 174105 (2011). DOI: http://dx.doi.org/10.1063/1.3656296 |
| 113. | Pu, M., Garrahan, J.P. & Hirst, J.D., Influence of solvent model on protein dynamics. Chem. Phys. Lett., 515, 283–289 (2011). DOI: http://dx.doi.org/10.1016/j.cplett.2011.09.026 |
| 112. | Do, H., Wheatley, R.J. & Hirst, J.D., Molecular simulation of the binary mixture of 1-1-1-2-tetrafluoroethane and carbon dioxide. Phys. Chem. Chem. Phys, 13, 15708–15713 (2011). DOI: http://dx.doi.org/10.1039/c1cp21419e |
| 111. | Baker, J.A., Hirst, J.D., Molecular Dynamics Simulations Using Graphics Processing Units. Mol. Inf., 30, 498–504 (2011). DOI: http://dx.doi.org/10.1002/minf.201100042 |
| 110. | Robinson, D., Besley, N.A., O'Shea, P. & Hirst, J.D., Water order profiles on phospholipid / cholesterol membrane bilayer surfaces. J. Comput. Chem., 32, 2613 (2011). DOI: http://dx.doi.org/10.1002/jcc.21840 |
| 109. | Gaigeot, M.-P., Besley, N.A. & Hirst, J.D, Modelling the infrared and circular dichroism spectroscopy of linear and cyclic diamides. J. Phys. Chem. B, 115, 5562–5535 (2011). DOI: http://dx.doi.org/10.1021/jp111140f |
| 108. | Robinson, D., Besley, N.A., O'Shea, P. & Hirst, J.D., Di-8-ANEPPS Emission Spectra in Phospholipid / Cholesterol Membranes: A Theoretical Study. J. Phys. Chem. B, 115, 4160–4167 (2011). DOI: http://dx.doi.org/10.1021/jp1111372 |
| 107. | Oakley, M.T., Do, H., Hirst, J.D. & Wheatley, R.J., First principles predictions of thermophysical properties of refrigerant mixtures. J. Chem. Phys, 134, 114518 (2011). DOI: http://dx.doi.org/10.1063/1.3567308 |
| 106. | Chen, P., Evans, C.-L., Hirst, J.D., Searle, M.S., Structural insights into the two sequential folding transition states of the PB1 domain of NBR1 from Φ value analysis and biased molecular dynamics simulations. Biochemistry, 50, 125–135 (2011). DOI: http://dx.doi.org/10.1021/bi1016793 |
| 105. | Turpin, E.R., Bonev, B.B., Hirst, J.D., Stereoselective disulfide formation stabilizes the local peptide conformation in Nisin mimics. Biochemistry, 49, 9594–9603 (2010). DOI: http://dx.doi.org/10.1021/bi101214t |
| 104. | Do, H., Wheatley, R.J. & Hirst, J.D., Microscopic structure of liquid 1-1-1-2-tetrafluoroethane (R134a) from Monte Carlo simulation. Phys. Chem. Chem. Phys., 12, 13266–13272 (2010). DOI: http://dx.doi.org/10.1039/C0CP00620C |
| 103. | Jiang, J., Abramavicius, D., Falvo, C., Bulheller, B.M., Hirst, J.D. & Mukamel, S., Simulation of two-dimensional ultraviolet spectroscopy of amyloid fibrils. J. Phys. Chem. B., 114, 12150–12156 (2010). DOI: http://dx.doi.org/10.1021/jp1046968 |
| 102. | Kountouris, P. & Hirst, J.D., Predicting β-turns and their types using predicted backbone dihedral angles and secondary structures. BMC Bioinformatics, 11, 407 (2010). DOI: http://dx.doi.org/10.1186/1471-2105-11-407 |
| 101. | Jain, P. & Hirst, J.D., Automatic structure classification of small proteins using random forest. BMC Bioinformatics, 11, 364 (2010). DOI: http://dx.doi.org/10.1186/1471-2105-11-364 |
| 100. | Jiang, J., Abramavicius, D., Bulheller, B.M., Hirst, J.D. & Mukamel, S., Ultraviolet spectroscopy of protein backbone transitions in aqueous solution: QM/MM simulations. J. Phys. Chem. B, 114, 8270–8277 (2010). DOI: http://dx.doi.org/10.1021/jp101980a |
| 99. | Abramavicius, D., Jiang, J., Bulheller, B.M., Hirst, J.D. & Mukamel, S., Simulation Study of Chiral Two-Dimensional Ultraviolet Spectroscopy of the Protein Backbone. J. Am. Chem. Soc., 132, 7769–7775 (2010). DOI: http://dx.doi.org/10.1021/ja101968g |
| 98. | Bromley, E.H.C., Channon, K.J., King, P.J.S., Mahmoud, Z.N., Banwell, E.F., Butler, M.F., Crump, M.P., Dafforn, T.R., Hicks, M.R., Hirst, J.D., Rodger, A. & Woolfson, D.N., The assembly pathway of a designed α-helical protein fiber. Biophys. J., 98, 1668–1676 (2010). DOI: http://dx.doi.org/10.1016/j.bpj.2009.12.4309 |
| 97. | Smith, R.E., Liang, M., Bacardit, J., Stout, M., Krasnogor, N. & Hirst, J.D., A Learning Classifier System with Mutual-Information-Based Fitness. Evolutionary Intelligence, 3, 31–50 (2010). DOI: http://dx.doi.org/10.1007/s12065-010-0037-9 |
| 96. | Do, H., Wheatley, R.J. & Hirst, J.D., Gibbs ensemble Monte Carlo simulations of binary mixtures of methane, difluoromethane and carbon dioxide. J. Phys. Chem. B, 114, 3879–3886 (2010). DOI: http://dx.doi.org/10.1021/jp909769c |
| 95. | Hussain, A., Melville, J.L. & Hirst, J.D., Molecular docking and QSAR of aplyronine A and analogues: potent inhibitors of actin. J. Comput.-Aided Mol. Des., 24, 42005 (2010). DOI: http://dx.doi.org/10.1007/s10822-009-9307-y |
| 94. | Spowage, B.M., Bruce, C.L. & Hirst, J.D., Interpretable correlation descriptors for quantitative structure-activity relationships. J. Cheminf, 1, 22 (2009). DOI: http://dx.doi.org/10.1186/1758-2946-1-22  |
| 93. | Kountouris, P. & Hirst, J.D., Prediction of backbone dihedral angles and protein secondary structure using support vector machines. BMC Bioinformatics, 10, 437 (2009). DOI: http://dx.doi.org/10.1186/1471-2105-10-437 |
| 92. | Robinson, D., Besley, N.A., O'Shea, P. & Hirst, J.D., Calculating the fluorescence of 5-hydroxytryptophan in proteins. J. Phys. Chem. B, 113, 14521–14528 (2009). DOI: http://dx.doi.org/10.1021/jp9071108 |
| 91. | Jain, P. & Hirst, J.D., Exploring protein structural dissimilarity to facilitate structure classification. BMC Structural Biology, 9, 60 (2009). DOI: http://dx.doi.org/10.1186/1472-6807-9-60 |
| 90. | Bulheller, B.M., Rodger, A., Hicks, M.R., Dafforn, T.R., Serpell, L.C., Marshall, K.E., Bromley, E.H.C., King, P.J.S., Channon, K.J., Woolfson, D.N. & Hirst, J.D., Flow linear dichroism of some prototypical proteins. J. Am. Chem. Soc., 131, 13305–13314 (2009). DOI: http://dx.doi.org/10.1021/ja902662e  |
| 89. | Bulheller, B.M., Pantoş G.D., Sanders, J.K.M. & Hirst, J.D., Electronic structure and circular dichroism spectroscopy of naphthalenediimide nanotubes. Phys. Chem. Chem. Phys., 11, 6060–6065 (2009). DOI: http://dx.doi.org/10.1039/b905187b  |
| 88. | Jain, P., Garibaldi, J.M. & Hirst, J.D., Supervised machine learning algorithms for protein structure classification. Comp. Biol. Chem., 33, 216–223 (2009). DOI: http://dx.doi.org/10.1016/j.compbiolchem.2009.04.004 |
| 87. | Melville, J.L., Burke, E.K. & Hirst, J.D., Machine Learning in Virtual Screening. Comb. Chem. & High Thr. Scr., 12, 332–343 (2009). DOI: http://dx.doi.org/10.2174/138620709788167980 |
| 86. | Robinson, D., Besley, N.A., Lunt, E.A.M., O'Shea, P. & Hirst, J.D., Electronic structure of 5-hydroxyindole: from gas-phase to explicit solvation. J. Phys. Chem. B, 113, 2535–2541 (2009). DOI: http://dx.doi.org/10.1021/jp808943d |
| 85. | Bulheller, B.M. & Hirst, J.D., DichroCalc–circular and linear dichroism online. Bioinformatics, 25, 539–540 (2009). DOI: http://dx.doi.org/10.1093/bioinformatics/btp016 |
| 84. | Bacardit, J., Stout, M., Hirst, J.D., Valencia, A., Smith R.E., & Krasnogor, N., Automated alphabet reduction for protein datasets. BMC Bioinformatics, 10, 6 (2009). DOI: http://dx.doi.org/10.1186/1471-2105-10-6 |
| 83. | Stout, M., Bacardit, J., Hirst, J.D., Smith, R.E. & Krasnogor, N., Prediction of Topological Contacts in Proteins Using Learning Classifier Systems. Soft Comput., 13, 245–258 (2009). DOI: http://dx.doi.org/10.1007/s00500-008-0318-8 |
| 82. | Hamby, S.E. & Hirst, J.D., Prediction of Glycosylation Sites Using Random Forests. BMC Bioinformatics, 9, 500 (2008). DOI: http://dx.doi.org/10.1186/1471-2105-9-500 |
| 81. | Oakley, M.T., Barthel, D., Bykov, Y., Garibaldi, J.M., Burke, E.K., Krasnogor, N. & Hirst, J.D., Search Strategies in Structural Bioinformatics. Curr. Prot. Peptide. Sci., 9, 260–274 (2008). DOI: http://dx.doi.org/10.2174/138920308784534032 |
| 80. | Stout, M., Bacardit, J., Hirst, J.D. & Krasnogor, N., Prediction of Recursive Convex Hull Class Assignments for Protein Residues Using Learning Classifier Systems. Bioinformatics, 24, 916–923 (2008). DOI: http://dx.doi.org/10.1093/bioinformatics/btn050 |
| 79. | Evans, C.-L., Long, J.E., Gallagher, T.R.A., Hirst, J.D. & Searle, M.S., Conformation and dynamics of the three-helix bundle UBA domain of p62 from experiment and simulation. Proteins: Structure, Function & Bioinformatics, 71, 227–240 (2008). DOI: http://dx.doi.org/10.1002/prot.21692 |
| 78. | Bulheller, B.M., Miles, A.J., Wallace, B.A. & Hirst, J.D., Charge-Transfer Transitions in the Vacuum-Ultraviolet of Protein Circular Dichroism Spectra. J. Phys. Chem. B, 112, 1866–1874 (2008). DOI: http://dx.doi.org/10.1021/jp077462k |
| 77. | Barthel, D., Hirst, J.D., Blazewicz, J., Burke, E.K. & Krasnogor, N., ProCKSI: a decision support system for Protein (Structure) Comparison, Knowledge, Similarity and Information. BMC Bioinformatics, 8, 416 (2007). DOI: http://dx.doi.org/10.1186/1471-2105-8-416 |
| 76. | Bacardit, J., Stout, M., Hirst, J.D., Sastry, K. Llora, X. & Krasnogor, N., Automated alphabet reduction method with evolutionary algorithms for protein structure prediction. GECCO '07: Proceedings of the 9th annual conference on Genetic and evolutionary computation, London, England, (ISBN 978-1-59593-697-4), 346–353 (2007). DOI: http://dx.doi.org/10.1145/1276958.1277033 |
| 75. | Melville, J.L., Moal, I.H., Baker-Glenn, C., Shaw, P.E, Pattenden, G. & Hirst, J.D., The Structural Determinants of Macrolide-Actin Binding: In Silico Insights. Biophys. J., 92, 3862–3867 (2007). DOI: http://dx.doi.org/10.1529/biophysj.106.103580 |
| 74. | Bulheller, B.M., Rodger, A. & Hirst, J.D., Circular and Linear Dichroism of Proteins. Phys. Chem. Chem. Phys., 9, 2020–2035 (2007). DOI: http://dx.doi.org/10.1039/b615870f  |
| 73. | Oakley, M.T., Guichard, G. & Hirst, J.D., Calculations on the Electronic Excited States of Ureas and Oligoureas. J. Phys. Chem. B, 111, 3274–3279 (2007). DOI: http://dx.doi.org/10.1021/jp067890a |
| 72. | Melville, J.L. & Hirst, J.D., TMACC: Interpretable Correlation Descriptors for Quantitative Structure-Activity Relationships. J. Chem. Inf. Mod., 47, 626–634 (2007). DOI: http://dx.doi.org/10.1021/ci6004178  |
| 71. | Vincent, E., Saxton, J., Baker-Glenn, C., Moal, I., Hirst, J.D., Pattenden, G. & Shaw, P.E., Disruption of actin dynamics and SRF-dependent gene regulation by the marine macrolide ulapualide A and its synthetic analogues. Cell. Mol. Life Sci., 64, 487–497 (2007). DOI: http://dx.doi.org/10.1007/s00018-007-6427-1 |
| 70. | Dryden, I.L., Hirst, J.D. & Melville, J.L., Statistical analysis of unlabelled points: comparing molecules in cheminformatics. Biometrics, 63, 237–251 (2007). DOI: http://dx.doi.org/10.1111/j.1541-0420.2006.00622.x |
| 69. | Bruce, C.L., Melville, J.L., Pickett, S.D & Hirst, J.D., Contemporary QSAR Classifiers Compared. J. Chem. Inf. Mod., 47, 219–227 (2007). DOI: http://dx.doi.org/10.1021/ci600332j  |
| 68. | Melville, J.L., Riley, J.F. & Hirst, J.D., Similarity by Compression. J. Chem. Inf. Mod., 47, 25–33 (2007). DOI: http://dx.doi.org/10.1021/ci600384z  |
| 67. | Oakley, M.T. & Hirst, J.D., Charge-Transfer Transitions in Protein Circular Dichroism Calculations. J. Am. Chem. Soc., 128, 12414–12415 (2006). DOI: http://dx.doi.org/10.1021/ja0644125  |
| 66. | Bacardit, J., Stout, M., Krasnogor, N., Hirst, J.D., & Blazewicz, J., Coordination number prediction using learning classifier systems: performance and interpretability. In: Proceedings of the 8th Annual Conference on Genetic and Evolutionary Computation (Seattle, Washington, USA, July 08 - 12, 2006). GECCO '06, ACM Press, New York, NY, 247–254 (2006). DOI: http://dx.doi.org/10.1145/1143997.1144041 |
| 65. | Rogers, D.M., Hirst, J.D., Lee, E.P.F. & Wright, T.G., Ab Initio Study of the Toluene Dimer. Chem. Phys. Lett., 427, 410–413 (2006). DOI: http://dx.doi.org/10.1016/j.cplett.2006.07.022  |
| 64. | Jansen, T.L.C., Dijkstra, A.G., Watson, T.M., Hirst, J.D. & Knoester, J., Modeling the amide I bands of small peptides. J. Chem. Phys., 125, 44312/1 – 44312/9 (2006). DOI: http://dx.doi.org/10.1063/1.2218516 |
| 63. | Stout, M., Bacardit, J., Hirst, J.D., Krasnogor, N., & Blazewicz, J., From HP Lattice Models to Real Proteins: coordination number prediction using Learning Classifier Systems. Lectures Notes in Computer Science, 3907, 208–220 (2006). DOI: http://dx.doi.org/10.1007/11732242_19 |
| 62. | Oakley, M.T., Bulheller, B.M. & Hirst, J.D., First Principles Calculations of Protein Circular Dichroism in the Far-Ultraviolet and Beyond. Chirality, 18, 340–347 (2006). DOI: http://dx.doi.org/10.1002/chir.20264 |
| 61. | Rogers, D.M., Besley, N.A., O'Shea, P. & Hirst, J.D., Modeling the Absorption Spectrum of Tryptophan in Proteins. J. Phys. Chem. B, 109, 23061–23069 (2005). DOI: http://dx.doi.org/10.1021/jp053309j |
| 60. | Blackburne, B.P. & Hirst, J.D., Population Dynamics Simulations of Functional Model Proteins. J. Chem. Phys., 123, 154907/1–154907/9 (2005). DOI: http://dx.doi.org/10.1063/1.2056545 |
| 59. | Oakley, M.T., Garibaldi, J.M. & Hirst, J.D., Lattice models of peptide aggregation: Evaluation of conformational search algorithms. J. Comp. Chem., 26, 1638–1646 (2005). DOI: http://dx.doi.org/10.1002/jcc.20306 |
| 58. | Melville, J.L., Lovelock, K.J.R., Wilson, C., Allbutt, B., Burke, E.K., Lygo, B. & Hirst, J.D., Exploring Phase-Transfer Catalysis with Molecular Dynamics and 3D/4D Quantitative Structure-Selectivity Relationships. J. Chem. Inf. Mod., 45, 971–981 (2005). DOI: http://dx.doi.org/10.1021/ci050051l |
| 57. | Watson, T.M. & Hirst, J.D., Theoretical Studies of the Amide I Vibrational Frequencies of [Leu]-enkephalin. Mol. Phys., 103, 1531–1546 (2005). DOI: http://dx.doi.org/10.1080/00268970500052387 |
| 56. | McNeany, T.J. & Hirst, J.D., Inhibition of the Tyrosine Kinase, Syk, Analyzed by Stepwise Non-Parametric Regression. J. Chem. Inf. Mod., 45, 768–776 (2005). DOI: http://dx.doi.org/10.1021/ci049631t |
| 55. | Wood, M.J. & Hirst, J.D., Protein Secondary Structure Prediction with Dihedral Angles. Proteins: Structure, Function & Bioinformatics, 59, 476–481 (2005). DOI: http://dx.doi.org/10.1002/prot.20435 |
| 54. | Pelta, D.A., Krasnogor, N., Bousono-Calzon, C., Verdegay, J.L., Hirst, J.D., Burke, E.K., A Fuzzy Sets based Generalization of Contact Maps for the Overlap of Protein Structures. Fuzzy Sets and Systems, 152, 103–121 (2005). DOI: http://dx.doi.org/10.1016/j.fss.2004.10.017 |
| 53. | Besley, N.A, Oakley, M.T., Cowan, A.J. & Hirst, J.D., A Sequential Molecular Mechanics/Quantum Mechanics Study of the Electronic Spectra of Amides. J. Am. Chem. Soc., 126, 13502–13511 (2004). DOI: http://dx.doi.org/10.1021/ja047603l  |
| 52. | Lygo, B., Andrews, B.I., Hirst, J.D., Melville, J.L., Peterson, J.A. & Slack, D., Rapid screening of cinchona alkaloid derived phase-transfer catalysts. Application in the optimization of a glycine imine alkylation. Chim Oggi, 22(9), 41920 (2004). DOI: http://dx.doi.org/10.1002/chin.200549258 |
| 51. | Rogers, D.M. & Hirst, J.D., First Principles Calculations of Protein Circular Dichroism in the Near-Ultraviolet. Biochemistry, 43, 11092–11102 (2004). DOI: http://dx.doi.org/10.1021/bi049031n  |
| 50. | Melville, J.L. & Hirst, J.D., On the stability of CoMFA models. J. Chem. Inf. Comput. Sci., 44, 1294–1300 (2004). DOI: http://dx.doi.org/10.1021/ci049944o |
| 49. | Melville, J.L., Andrews, B.I., Lygo, B. & Hirst, J.D., Computational screening of combinatorial catalyst libraries. Chem. Comm., 12, 1410–1411 (2004). DOI: http://dx.doi.org/10.1039/b402378a  |
| 48. | Watson, T.M. & Hirst, J.D., Vibrational Analysis of Capped [Leu]Enkephalin. Phys. Chem. Chem. Phys., 6, 2580–2587 (2004). DOI: http://dx.doi.org/10.1039/b315501c |
| 47. | Gilbert, A.T.B. & Hirst, J.D., Charge-Transfer Transitions in Protein Circular Dichroism Spectra. J. Mol. Struct. (THEOCHEM), 675, 53–60 (2004). DOI: http://dx.doi.org/10.1016/j.theochem.2003.12.038 |
| 46. | Rogers, D.M. & Hirst, J.D., Calculations of Protein Circular Dichroism from First Principles. Chirality, 16, 234–243 (2004). DOI: http://dx.doi.org/10.1002/chir.20018 |
| 45. | Watson, T.M. & Hirst, J.D., Calculating Vibrational Frequencies of Amides: from Formamide to Concanavalin A. Phys. Chem. Chem. Phys., 6, 998–1005 (2004). DOI: http://dx.doi.org/10.1039/b312181j |
| 44. | Wood, M.J. & Hirst, J.D., Predicting Protein Secondary Structure by Cascade- Correlating Neural Networks. Bioinformatics, 20, 419–420 (2004). DOI: http://dx.doi.org/10.1093/bioinformatics/btg423 |
| 43. | Rogers, D.M. & Hirst, J.D., Ab Initio Studies of Aromatic Side-Chains in Gas Phase and Solution. J. Phys. Chem. A, 107, 11191–11200 (2003). DOI: http://dx.doi.org/10.1021/jp036081d |
| 42. | Hirst, J.D., Colella, K. & Gilbert, A.T.B., Electronic Circular Dichroism Spectra of Proteins from First Principles Calculations. J. Phys. Chem. B, 107, 11813–11819 (2003). DOI: http://dx.doi.org/10.1021/jp035775j |
| 41. | Watson, T.M. & Hirst, J.D., Influence of Electrostatic Environment on the Vibrational Frequencies of Proteins. J. Phys. Chem. A, 107, 6843–6849 (2003). DOI: http://dx.doi.org/10.1021/jp0344500 |
| 40. | Bhattacharjee, S., Tóth, G., Lovas, S. & Hirst, J.D., Influence of Tyrosine on the Electronic Circular Dichroism of Helical Peptides. J. Phys. Chem. B, 107, 8682–8688 (2003). DOI: http://dx.doi.org/10.1021/jp034517j |
| 39. | Blackburne, B.P. & Hirst, J.D., Three Dimensional Functional Model Proteins: Structure, Function and Evolution. J. Chem. Phys., 119, 3453–3460 (2003). DOI: http://dx.doi.org/10.1063/1.1590310 |
| 38. | Cox, K., Watson, T., Soultanas, P. & Hirst, J.D., Molecular Dynamics Simulations of a Helicase. Proteins: Structure, Function & Genetics, 52, 254–262 (2003). DOI: http://dx.doi.org/10.1002/prot.10400 |
| 37. | Hirst, J.D., Bhattacharjee, S. & Onufriev, A.V., Theoretical Studies of Time-Resolved Protein Folding. Faraday Discussions, 122, 253–267 (2003). DOI: http://dx.doi.org/10.1039/b200714b |
| 36. | Andrew, C.D., Bhattacharjee, S., Kokkoni, N., Hirst, J.D., Jones, G.R. & Doig, A.J., Stabilizing Interactions between Aromatic and Basic Side Chains in α-Helical Peptides. Tyrosine Effects on Helix Circular Dichroism. J. Am. Chem. Soc., 124, 12706–12714 (2002). DOI: http://dx.doi.org/10.1021/ja027629h  |
| 35. | Krasnogor, N., Blackburne, B.P., Burke, E.K. & Hirst, J.D., Multimeme Algorithms for Protein Structure Prediction. Proceedings of the 7th International Conference on Parallel Problem Solving from Nature, Granada, Spain, Publishers: Springer, pp 769–778 (2002). |
| 34. | Carr, B., Hart, W.E., Hirst, J.D., Krasnogor, N., Burke, E.K & Smith, J., Alignment of Protein Structures with a Memetic Evolutionary Algorithm. Proceedings of the Genetic and Evolutionary Computation Conference 2002, NewYork, USA, Publishers: Morgan Kaufmann, pp 1027–1034 (2002). |
| 33. | Watson, T.M. & Hirst, J.D., DFT Vibrational Frequencies of Amides and Amide Dimers. J. Phys. Chem. A, 106, 7858–7867 (2002). DOI: http://dx.doi.org/10.1021/jp025551l |
| 32. | Rodger, A., Rajendra, J., Mortimer, R., Andrews, T., Hirst, J.D., Gilbert, A.T.B., Marrington, R., Dafforn, T.R., Hasall, D.J., Ardhammar, M., Nordén, B., Woolhead, C.A., Robinson, C., Pinheiro, T., Kazlauskaite, J., Seymour, M., Perez, N. & Hannon, M.J., Flow Oriented Linear Dichroism to Probe Protein Orientation in Membrane Environments. Phys. Chem. Chem. Phys., 4, 4051–4057 (2002). DOI: http://dx.doi.org/10.1039/b205080n |
| 31. | Hirst, J.D., McNeany, T.J., Howe, T. & Whitehead, L., Application of Non- Parametric Regression to Quantitative Structure-Activity Relationships. Bioorg. Med. Chem., 10, 1037–1041 (2002). DOI: http://dx.doi.org/10.1016/S0968-0896(01)00359-5  |
| 30. | Dang, Z. & Hirst, J.D., Short Hydrogen Bonds, Circular Dichroism and Over- Estimates of Peptide Helicity. Angew. Chemie Intl. Ed., 40, 3619–3621 (2001). DOI: http://dx.doi.org/10.1002/1521-3773(20011001)40:19<3619::AID-ANIE3619>3.0.CO;2-4 |
| 29. | Blackburne, B.P. & Hirst, J.D., Evolution of Functional Model Proteins. J. Chem. Phys., 115, 1935–1942 (2001). DOI: http://dx.doi.org/10.1063/1.1383051 |
| 28. | Besley, N.A., Brienne, M.-J. & Hirst, J.D., Electronic Structure of a Rigid Cyclic Diamide. J. Phys. Chem. B, 104, 12371–12377 (2000). DOI: http://dx.doi.org/10.1021/jp0024524 |
| 27. | Besley, N.A. & Hirst, J.D., Hydrogen Bonding in Protein Circular Dichroism Calculations. J. Mol. Struct. (THEOCHEM), 506, 161–167 (2000). DOI: http://dx.doi.org/10.1016/S0166-1280(00)00409-7 |
| 26. | Constans, P & Hirst, J.D., Non-Parametric Regressors Applied to Quantitative Structure-Activity Relationships. J. Chem. Inf. Comput. Sci., 40, 452–459 (2000). DOI: http://dx.doi.org/10.1021/ci990082e |
| 25. | Besley, N.A. & Hirst, J.D., Theoretical Studies toward Quantitative Protein Circular Dichroism Calculations. J. Am. Chem. Soc., 121, 9636–9644 (1999). DOI: http://dx.doi.org/10.1021/ja990627l |
| 24. | Besley, N.A. & Hirst, J.D., Ab Initio Study of the Electronic Spectrum of Formamide with Explicit Solvent. J. Am. Chem. Soc., 121, 8559–8566 (1999). DOI: http://dx.doi.org/10.1021/ja990064d |
| 23. | Hirst, J.D., The Evolutionary Landscapes of Functional Model Proteins. Protein Engineering, 12, 721–726 (1999). |
| 22. | Hirst, J.D. & Besley, N.A., Response to »Comment on 'Improving Protein Circular Dichroism Calculations in the Far-Ultraviolet through Reparametrizing the Amide Chromophore'«. J. Chem. Phys. [J. Chem. Phys. 109, 782-788 (1998)], 111, 2846–2847 (1999). DOI: http://dx.doi.org/10.1063/1.479563 |
| 21. | Hirst, J.D., Dominy, B., Guo, Z., Vieth, M. & Brooks III, C.L., Conformational and Energetic Aspects of Receptor-Ligand Recognition. Am. Chem. Soc. Symp. Series, 719, 13485 (1999). |
| 20. | Besley, N.A. & Hirst, J.D., Ab Initio Study of the Effect of Solvation on the Electronic Spectra of Formamide and N-Methylacetamide. J. Phys. Chem. A, 102, 10791–10797 (1998). DOI: http://dx.doi.org/10.1021/jp982645f |
| 19. | Hirst, J.D., Predicting Ligand Binding Energies. Curr. Opin. Drug Discovery & Development, 1, 28–33 (1998). |
| 18. | Hirst, J.D., Improving Protein Circular Dichroism Calculations through Better Ab Initio Models of the Amide Chromophore. Enantiomer, 3, 215–220 (1998). |
| 17. | Vieth, M., Hirst, J.D., Dominy, B.N., Daigler, H. & Brooks III, C.L., Assessing Search Strategies for Flexible Docking. J. Comp. Chem., 19, 1623–1631 (1998). DOI: http://dx.doi.org/10.1002/(SICI)1096-987X(19981115)19:14<1623::AID-JCC8>3.0.CO;2-L |
| 16. | Vieth, M., Hirst, J.D., Kolinski, A. & Brooks III, C.L., Assessing Energy Functions for Flexible Docking. J. Comp. Chem., 19, 1612–1622 (1998). DOI: http://dx.doi.org/10.1002/(SICI)1096-987X(19981115)19:14<1612::AID-JCC7>3.0.CO;2-M |
| 15. | Hirst, J.D., Improving Protein Circular Dichroism Calculations in the Far-Ultraviolet through Reparametrizing the Amide Chromophore. J. Chem. Phys., 109, 782–788 (1998). DOI: http://dx.doi.org/10.1063/1.476617 |
| 14. | Vieth, M., Hirst, J.D. & Brooks III, C.L., Do Active Site Conformations of Small Ligands Correspond to Low Free Energy Structures? J. Comput.-Aided Mol. Des., 12, 563–572 (1998). DOI: http://dx.doi.org/10.1023/A:1008055202136 |
| 13. | Hirst, J.D. & Persson, B.J., Ab Initio Calculations of the Vibrational and Electronic Spectra of Diketopiperazine. J. Phys. Chem. A, 102, 7519–7524 (1998). DOI: http://dx.doi.org/10.1021/jp982423h |
| 12. | Hirst, J.D., Hirst, D.M. & Brooks III, C.L., Multireference Configuration Interaction Calculations of Electronic States of N-Methylformamide, Acetamide, and N-Methylacetamide. J. Phys. Chem. A, 101, 4821–4827 (1997). DOI: http://dx.doi.org/10.1021/jp970675x |
| 11. | Hirst, J.D., Nonlinear Quantitative Structure-Activity Relationship for the Inhibition of Dihydrofolate Reductase by Pyrimidines. J. Med. Chem., 39, 3526–3532 (1996). DOI: http://dx.doi.org/10.1021/jm960197z |
| 10. | Hirst, J.D., Hirst, D.M. & Brooks III, C.L., Ab Initio Calculations of the Excited States of Formamide. J. Phys. Chem., 100, 13487–13491 (1996). DOI: http://dx.doi.org/10.1021/jp960597y |
| 9. | Hirst, J.D., Vieth, M., Skolnick, J. & Brooks III, C.L., Predicting Leucine Zipper Structures from Sequence. Protein Engineering, 9, 657–662 (1996). DOI: http://dx.doi.org/10.1093/protein/9.8.657 |
| 8. | Hirst, J.D. & Brooks III, C.L., Molecular Dynamics Simulations of Isolated Helices of Myoglobin. Biochemistry, 34, 7614–7621 (1995). DOI: http://dx.doi.org/10.1021/bi00023a007 |
| 7. | King, R.D., Hirst, J.D. & Sternberg, M.J.E., Drug Design by Machine Learning: A Comparative Study. Applications in Artificial Intelligence, 9, 213–233 (1995). |
| 6. | Hirst, J.D. & Brooks III, C.L., Helicity, Circular Dichroism and Molecular Dynamics of Proteins. J. Mol. Biol., 243, 173–178 (1994). DOI: http://dx.doi.org/10.1006/jmbi.1994.1644 |
| 5. | Hirst, J.D., King, R.D. & Sternberg, M.J.E., Quantitative Structure-Activity Relationships by Neural Networks and Inductive Logic Programming II. The Inhibition of Dihydrofolate Reductase by Triazines. J. Comput.-Aided Mol. Des., 8, 421–432 (1994). DOI: http://dx.doi.org/10.1007/BF00125376 |
| 4. | Hirst, J.D., King, R.D. & Sternberg, M.J.E., Quantitative Structure-Activity Relationships by Neural Networks and Inductive Logic Programming I. The Inhibition of Dihydrofolate Reductase by Pyrimidines. J. Comput.-Aided Mol. Des., 8, 405–420 (1994). DOI: http://dx.doi.org/10.1007/BF00125375 |
| 3. | King, R.D., Hirst, J.D. & Sternberg, M.J.E., New Approaches to QSAR: Neural Networks and Machine Learning. Perspectives in Drug Discovery and Design, 1, 279–290 (1993). DOI: http://dx.doi.org/10.1007/BF02174529 |
| 2. | Hirst, J.D. & Sternberg, M.J.E., Prediction of Structural and Functional Features of Protein and Nucleic Acid Sequences by Artificial Neural Networks. Biochemistry, 31, 7211–7218 (1992). DOI: http://dx.doi.org/10.1021/bi00147a001 |
| 1. | Hirst, J.D. & Sternberg, M.J.E., Prediction of ATP-Binding Motifs: A Comparison of a Perceptron-Type Neural Network and a Consensus Sequence Method. Protein Engineering, 4, 615–623 (1991). DOI: http://dx.doi.org/10.1093/protein/4.6.615 |


